- Sequencing Service
- Bioinformatics Service
- Sample Submission
- Price
- Contact
- Resources
- About
- Careers
- A-Z
- Webmail
- Inside the University Secured Page
-
Search UTHealth Houston
The Visium platform from 10x Genomics combines histology, protein detection, and spatially resolved whole transcriptome gene expression profiling to localize and quantify gene expression in the tissue context. Visium CytAssist assay offers Visium Spatial Gene Expression for FFPE, which is compatible with human and mouse formalin-fixed paraffin-embedded (FFPE) tissue sections. This assay utilizes RNA-templated ligation (RTL) of pairs of gene target probes for highly specific and sensitive detection of the whole transcriptome.
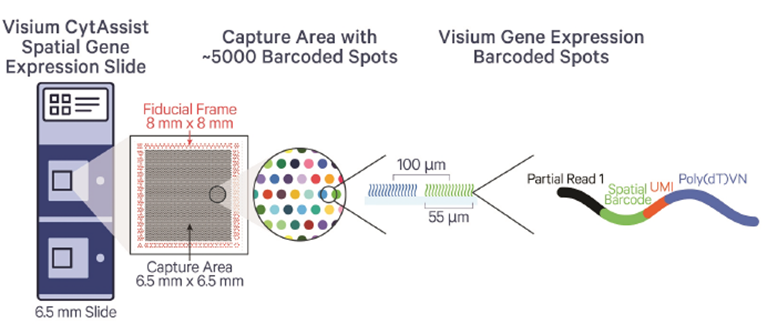
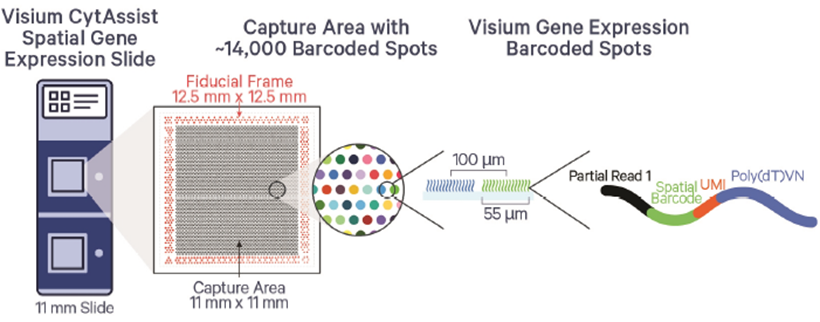
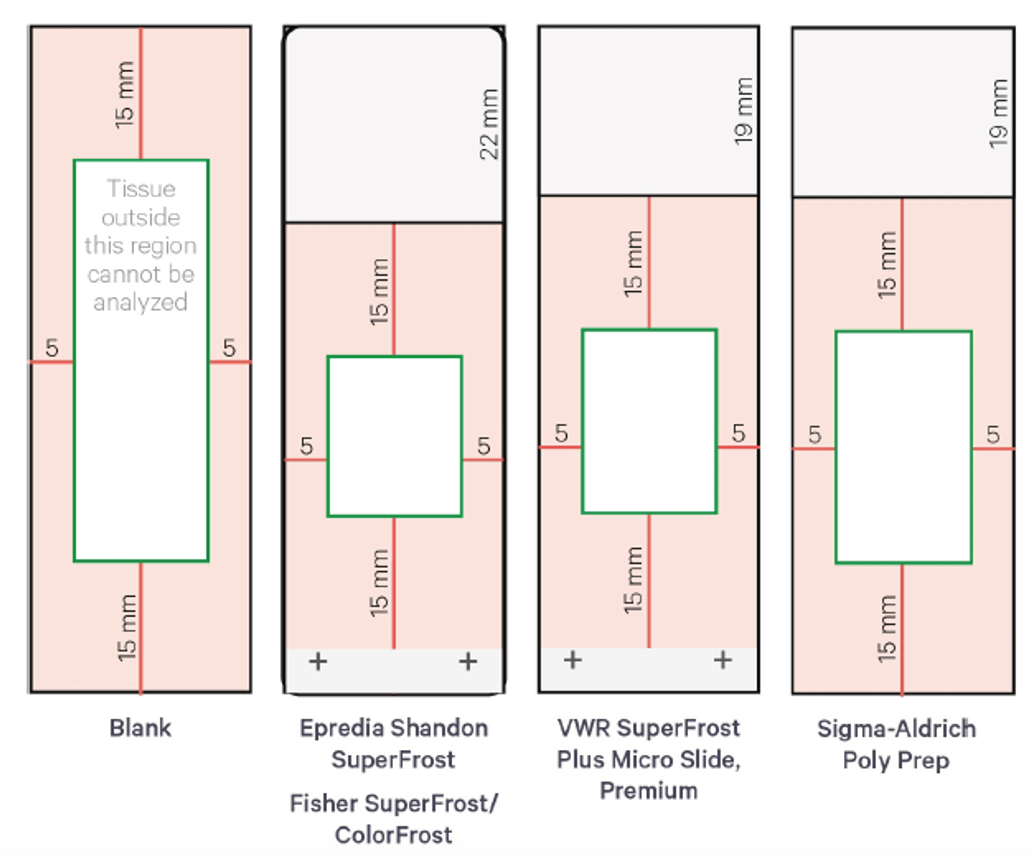
We only accept the slides after staining and imaging. The sample must locate inside the green frame of the example slides as in the figure below. We highly recommend to check the sample RIN if it is possible. The samples with DV200>30% are recommended for the assay. Please contact the core ([email protected] or 713-500-7933) for the detail information.
UTHealth B.R.A.I.N.S. HumanBiorepository and Histopathology Core or Research Histology Core Laboratory (RHCL) can help with the sample slide preparation.
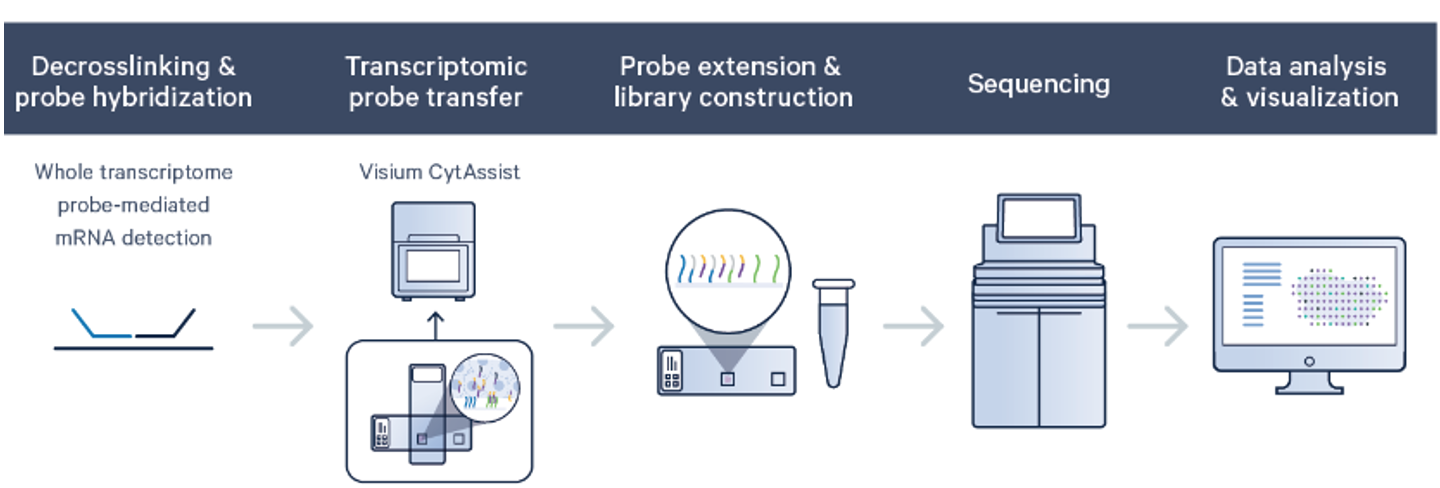
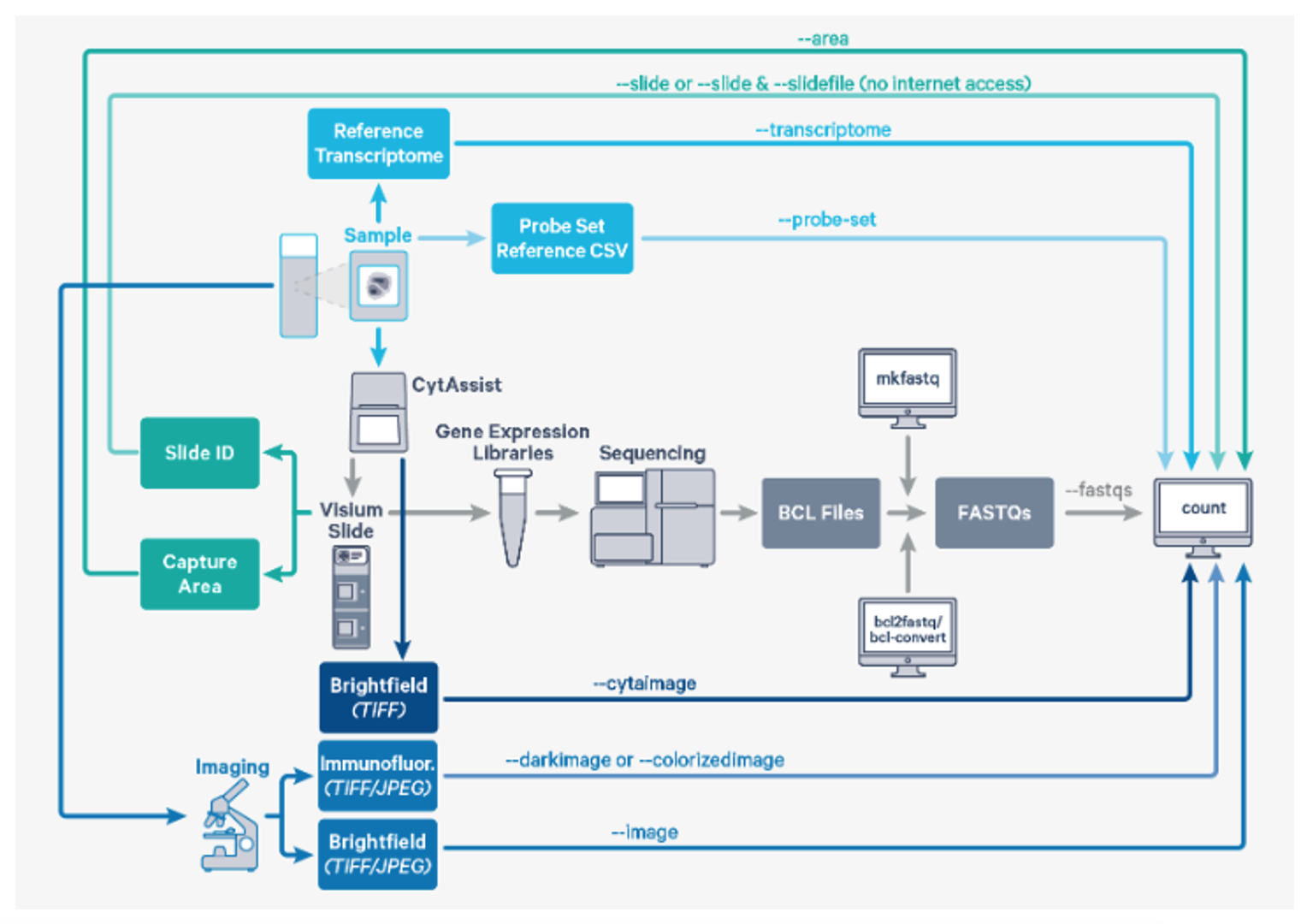
Space Ranger's pipelines analyze sequencing data produced from Visium Spatial Gene Expression for Formalin Fixed Paraffin Embedded (FFPE) and Fresh Frozen (FF) tissue samples. The CytAssist instrument aids in the transfer of analytes from the FFPE tissue section on a standard glass slides to the CytAssist Spatial Gene Expression Slide. The workflow diagram shows all the required inputs and the corresponding spaceranger flags for analyzing CytAssist enabled FFPE tissue samples: